See our publications
Algorithm and tool development for microbiome studies
Microbes are everywhere and they play important roles in sustaining life. Thanks to the development of sequencing technologies and others, microbiome studies have produced massive metagenomic data, and more recently other meta-omics including metatranscriptomic and metaproteomic data associated with different ecosystems, habitats and hosts, revealing insights into the composition, function and regulatory characteristics of the microbial communities. Analyzing microbiome data is computationally demanding, and still remains challenging.- RAPSearch & RAPSearch2 for fast protein similarity searches (our website; SourceForge)
- FragGeneScan for gene prediction in short reads, contigs, or even complete genomes; and sister program TransGeneScan for finding genes in metatranscriptomic sequences. (FragGeneScan at sourceforge; install FragGeneScan via anaconda)
- Pathway reconstruction for metagenomes (MinPath & MetaNetSam)
- Functional annotation for metagenomics (Fun4Me: all in one package)
- Check here to see more tools
Understanding the CRISPR–Cas systems and their applications (tools developed)
- The CRISPR–Cas adaptive immune system is an important defense system in bacteria and archaea, providing targeted defense against invasions of foreign nucleic acids (including viruses). The CRISPR (clusters of regularly interspaced short palindromic repeats) loci and cas (CRISPR-associated) genes are the two components of CRISPR–Cas immune systems: segments of invading DNAs are incorporated into host genomes in the CRISPR loci (forming spacers between repeats in CRISPR arrays), while cas genes encode Cas proteins that mediate the defense process.
- We have developed several computational approaches for the discovery and characterization of the CRISPR–Cas systems from metagenomic sequences (see Fig. 1). One exciting work that we have been working on is to apply the identified CRISPR-Cas systems to discover new invaders, and to study the arms race between the bacteria and the invaders (through the CRISPR-Cas immune systems). Check out our recent publications for more details.
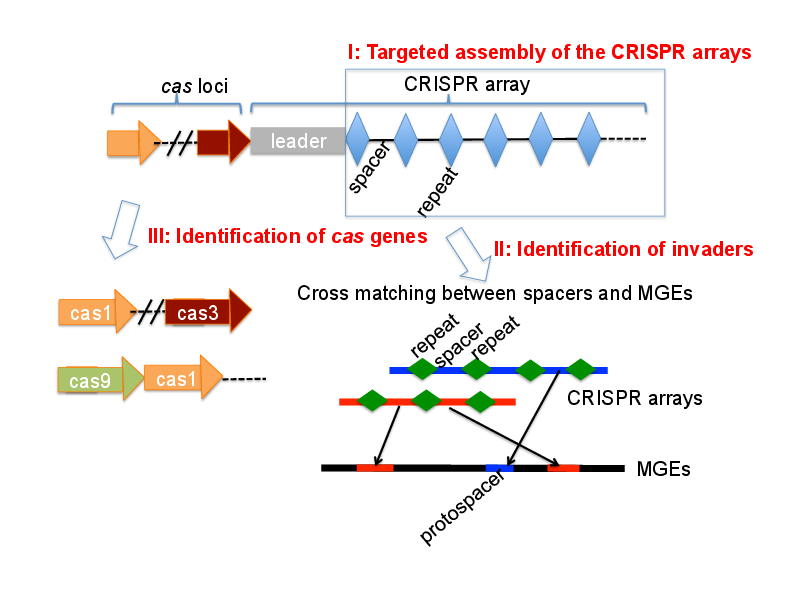
Fig. 1 Our work on the bacterial CRISPR-Cas immune system.
Protein sequence-structure-function relationship
- Protein structure prediction
- Methodology development
- Apply to human disease related proteins
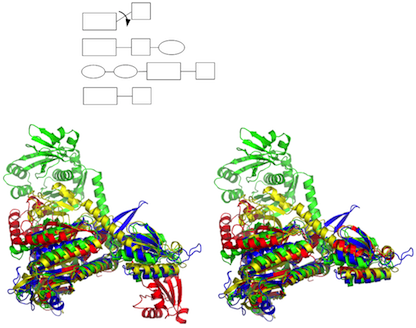
- Comparison of protein structures FATCAT and POSA are programs for protein structure comparison that consider the flexibility of protein structures (see Fig. 2).
- FATCAT (pairwise comparison allowing structural flexibility) (FATCAT server)
- POSA (multiple structure alignments using partial order graph representation) (POSA server)
- Protein design
- Tool development
- Applications: e.g., design specific inhibitors by interface redesign
Biological network
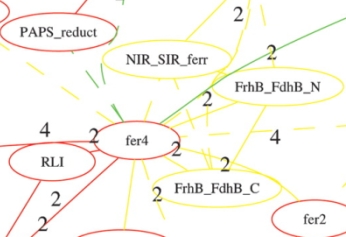
- Protein domain organization analysis (Fig 3); go to CADO server for details
- Biochemical pathway analysis
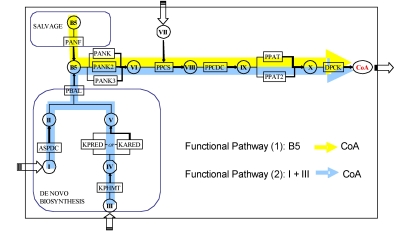
- Pathway variant detection (Fig 4)

 Yuzhen Ye & her lab
Yuzhen Ye & her lab